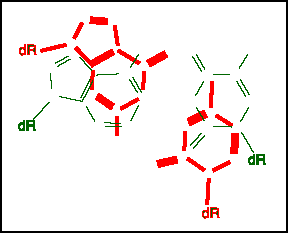
Stacked G:C and A:T base pairs
This figure looks down the axis of the helix at a G:C pair (red) over an A:T pair (green). The top pair is twisted 34 degrees relative to the pair below. The glycosidic bonds to the sugars are labeled "dR".
The is considerable overlap of the bases, but due to the twist it is not complete . As you can guess, the extent of over lap depends on the particular nucleotide sequence.
Since both sugars, and thus phosphates, are linked to the base pairs on the same side of the helix axis, the two phosphodiester chains are closer together at the lower left (the minor grove) than they are at the upper right (the major grove).