|
Determination of the relative amount of radioactivity in the phosphates is incredibly time consuming
(you don't want to know the details). We can only afford a few
time points, but we want to know the relative radioactivity over
the complete time span of the experiment.
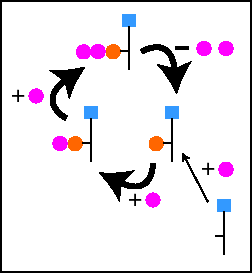
A pool model is the solution. In pharmacokinetic models a pool is a physical compartment, and
transport is flow between compartments. In the radioactive labeling
case a pool is a single species of nucleotide, and transport between
pools represents the chemical reaction that converts one form
to the other. The math is the same.
The raw kinetics of isotope incorporation (counts per minute, cpm) into interior
(orange) and terminal (red) nucleotides, top panel, left figure,
are nonlinear, with different shapes for each. The ratio between
interior and terminal thus changes with time after isotope addition,
about 10 percent at 20 seconds, and 0.2 percent at 5 minutes.
We can convert to absolute amounts, e.g. nanomoles (n moles) knowing the relative activities of
the nucleotides. Now the two curves, in the lower panel, are linear
with time. This just indicates that RNA is being made at a constant
rate. The ratio is also a constant, indicating an average length
of about 6000 nucleotides. |